This post is an update to the previous one on working with HDF-EOS files in R. There’s a neat little function in the MODIS package called runGdal that allows, with just a few lines of code, to a) download MODIS data over particular vertical and horizontal coordinates b) extract the desired layer(s) from the HDF-EOS file c) reproject and and mosaic your data, and d) save your imagery in geotiff format.
# install the package
install.packages("MODIS", repos="http://R-Forge.R-project.org")
# load it
require(MODIS)
# set the GDAL directory. In my case, GDAL is installed with QGIS
MODISoptions(gdalPath="C:/Program Files/QGIS Lyon/bin/")
# set the time period for which you want the imagery
dates1 <- as.POSIXct( as.Date(c("01/03/2015","30/06/2015"),format = "%d/%m/%Y") ) # specify your beginning and end dates
dates2 <- transDate(dates1[1],dates1[2]) # transform them into a readable format
# manually select the MODIS product, bands, and the horizontal and vertical location of the desired MODIS tile.
product <- "MOD13Q1" # MODIS vegetation indices
bands <- "011" # here I select the 2nd (EVI) and 3rd (QC flags) bands of the HDF file
# set your tile horizontal values, you can select multiple rows, they will be mosaicked together
h = c("20","21")
# set your tile vertical values, you can select multiple rows, they will be mosaicked together
v = c("04","05")
# proceed to download HDFs, project, and mosaic them
runGdal(product=product,begin=dates2$beginDOY,end = dates2$endDOY,tileH = h,tileV = v, resamplingType="bilinear", SDSstring = bands, outProj="4326")
# The default location is in the ~home_folder/MODIS_ARC/PROCESSED
# After the download is complete you can stack the processed TIFS
evi.list = list.files(path="C:/Users/Hakim/Documents/MODIS_ARC/PROCESSED/MOD13Q1.006_20160326155400/", full.names = T, pattern="*16_days_EVI.tif$")
evi.stack = stack(lapply(evi.list, raster))Once the runGDal function is executed, the data will begin to download. The HDF files will be stored in ~home_folder/MODIS_ARC/MODIS. This process can take a while depending on the duration of your study, the speed of your internet connection, the number of layers you selected, and the temporal interval of your layers.
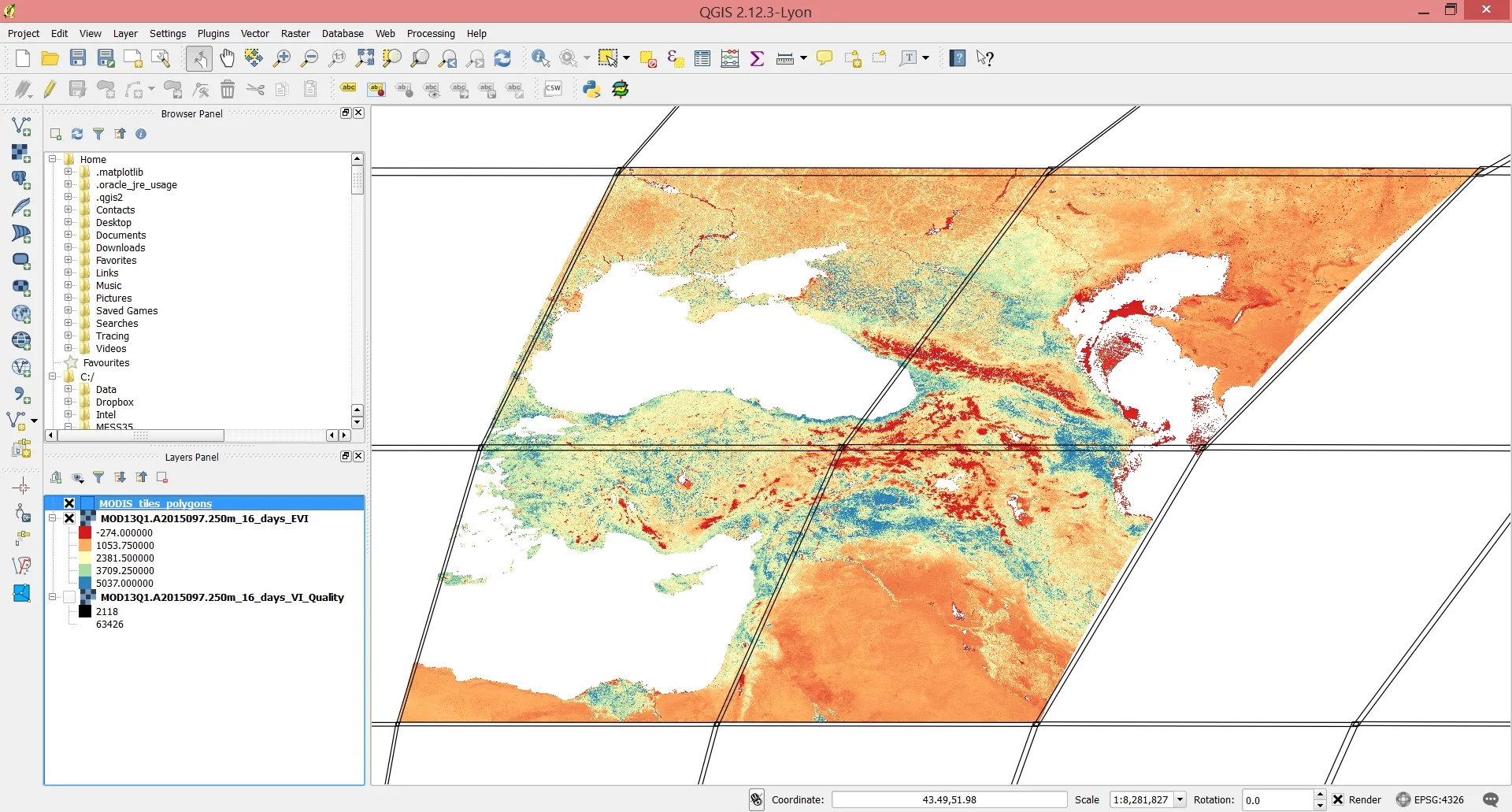
The output of the reprojection and mosaicking operations will be stored in ~home_folder/MODIS_ARC/PROCESSED. Each layer you select will be mosaicke (or not) according to the number of MODIS coordinates you select. For example, had we selected just h21 and v05, we would only have the lower right tile in the image below and no mmosaicking would be necessary. But because we selected two vertical and two horizontal values, they need to be mosaicked together in order to get a continuous satellite image of the study area.
Screenshot of the final mosaicked imagery loaded into QGIS along with the boundaries of the MODIS tiles.